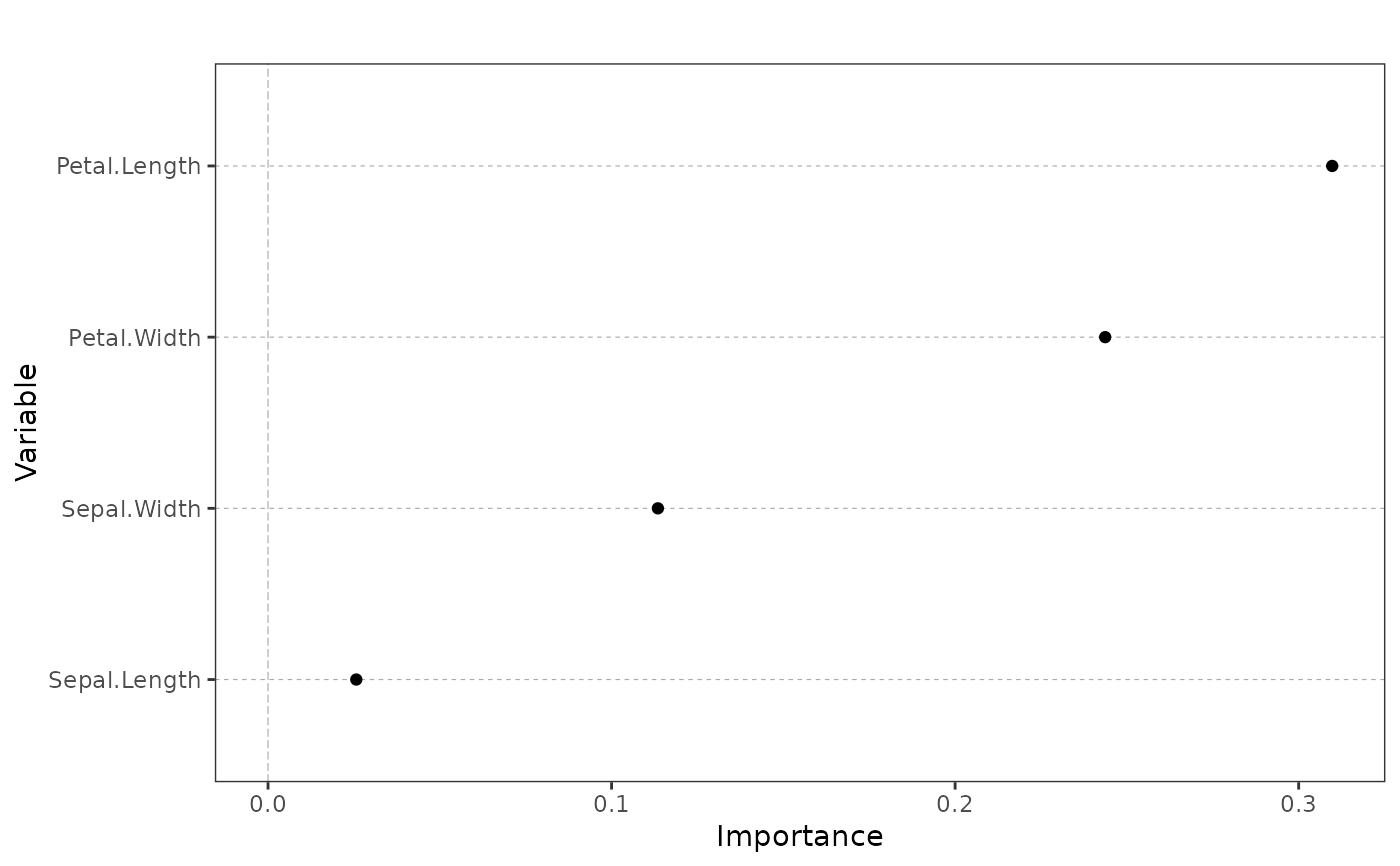
Dot plot of variable importance
ggVarImp.RdPlots the importance of the covariates of a supervised learning model in a dot plot.
ggVarImp(importance, sort=TRUE, xlabel="Importance", ylabel="Variable", main="")Arguments
- importance
numeric vector. The vector of the importances of the covariates. Should be a named vector.
- sort
logical. Whether the vector of importances should be sorted or not. Default is TRUE.
- xlabel
character. Title of the x axis.
- ylabel
character. Title of the y axis.
- main
character. Title of the plot.
See also
varImp,varImpAUC,fastvarImp,fastvarImpAUC
Examples
data(iris)
iris2 = iris
iris2$Species = factor(iris$Species == "versicolor")
iris.cf = party::cforest(Species ~ ., data = iris2,
control = party::cforest_unbiased(mtry = 2, ntree = 50))
imp <- fastvarImpAUC(object = iris.cf, parallel = FALSE)
ggVarImp(imp)