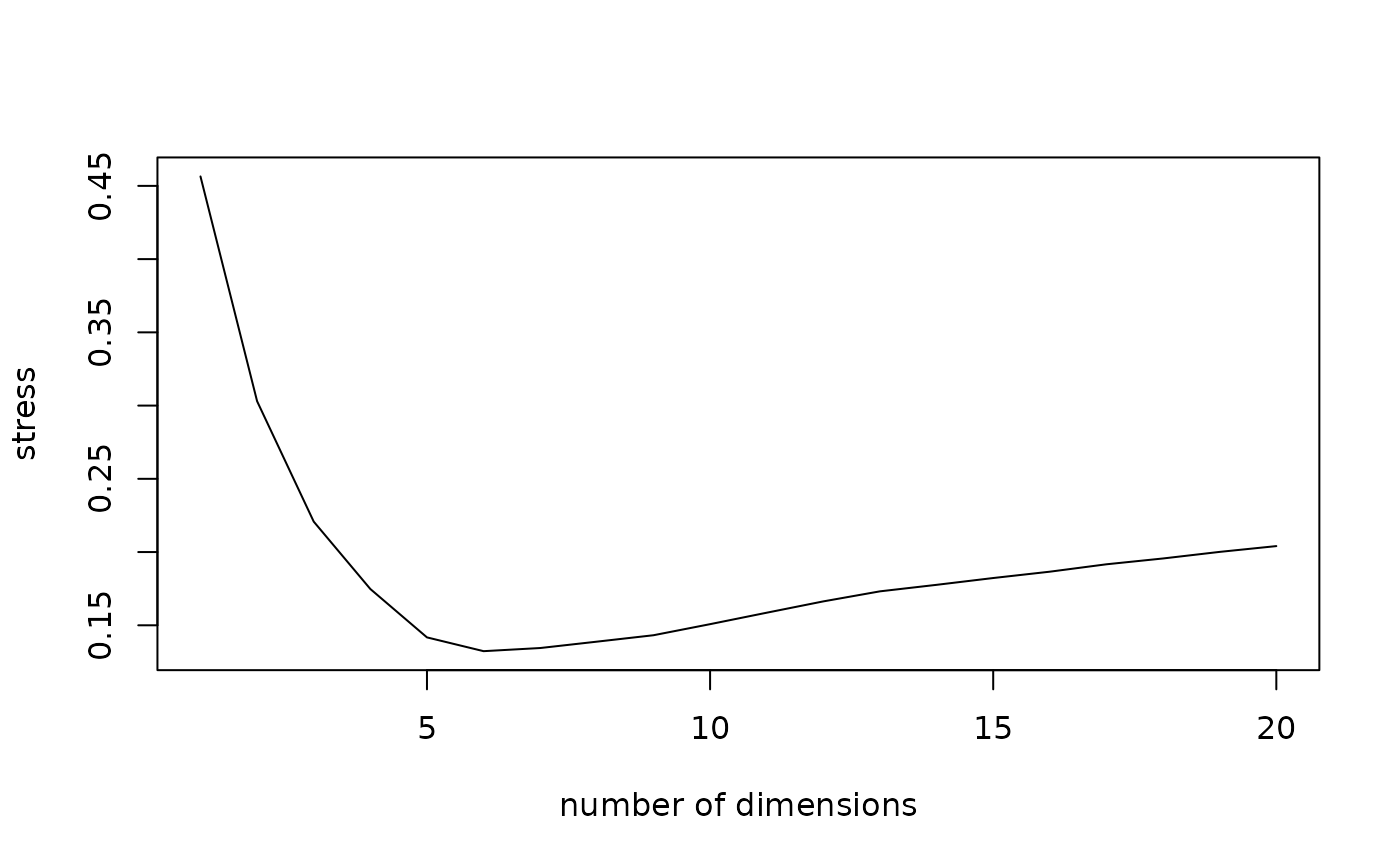
Stress measure of multidimensional scaling factors
seqmds.stress.RdComputes stress measure of multidimensional scaling data for different number of dimensions of the represented space
seqmds.stress(seqdist, mds)Arguments
- seqdist
a dissimilarity matrix or a dist object (see
dist)- mds
a matrix with coordinates in the represented space (dimension 1 in column 1, dimension 2 in column 2, etc.)
Value
A numerical vector of stress values.
References
Piccarreta R., Lior O. (2010). Exploring sequences: a graphical tool based on multi-dimensional scaling, Journal of the Royal Statistical Society (Series A), Vol. 173(1), 165-184.
Examples
data(trajact)
seqact <- seqdef(trajact)
#> [>] 6 distinct states appear in the data:
#> 1 = 1
#> 2 = 2
#> 3 = 3
#> 4 = 4
#> 5 = 5
#> 6 = 6
#> [>] state coding:
#> [alphabet] [label] [long label]
#> 1 1 1 1
#> 2 2 2 2
#> 3 3 3 3
#> 4 4 4 4
#> 5 5 5 5
#> 6 6 6 6
#> [>] 500 sequences in the data set
#> [>] min/max sequence length: 37/37
dissim <- seqdist(seqact, method="HAM")
#> [>] 500 sequences with 6 distinct states
#> [>] creating a 'sm' with a single substitution cost of 1
#> [>] creating 6x6 substitution-cost matrix using 1 as constant value
#> [>] 377 distinct sequences
#> [>] min/max sequence lengths: 37/37
#> [>] computing distances using the HAM metric
#> [>] elapsed time: 0.114 secs
mds <- cmdscale(dissim, k=20, eig=TRUE)
stress <- seqmds.stress(dissim, mds)
plot(stress, type='l', xlab='number of dimensions', ylab='stress')